
J. Adv. Vet. Anim. Res., Volume 3 Issue 4, December 2016, Pages 368-374.
DOI: http://doi.org/10.5455/javar.2016.c175
Comparison of different DNA isolation methods and use of dodecyle trimethyl ammonium bromide (DTAB) for the isolation of DNA from meat products
Yusuf Özşensoy and Seyda Şahin
Affiliations:Yusuf Özşensoy: Department of Veterinary Biometrics and Genetics, Faculty of Veterinary Medicine, Cumhuriyet University, 58140, Sivas, Turkey.
Seyda Şahin: Department of Food Hygiene and Technology, Faculty of Veterinary Medicine, Cumhuriyet University, 58140, Sivas, Turkey.
Correspondence: Yusuf Özşensoy
Received: Sep 19, 2016 Revised: Oct 27, 2016 Accepted: Nov 13, 2016 Published Online: Nov 13, 2016
How to cite: Özşensoy Y, Şahin S (2016). Comparison of different DNA isolation methods and use of dodecyle trimethyl ammonium bromide (DTAB) for the isolation of DNA from meat products. Journal of Advanced Veterinary and Animal Research, 3(4): 368-374.
ABSTRACT
Objective:The
identification of meat species in meat products is important for
protection of human health,economic reasons, religious factors and for
controlling the compliance with food regulations. For
this purpose, DNA must be obtained in good
quality and quantity. The aim of this study was
to compare different DNA isolation methods from different meat products.
Materials and
methods: Comparison
among different DNA
isolation methods was done. DNA was
isolated from different meat products (e.g.,
sucuk, salami, sausage, braised meet, meatball and pastrami). The
methods
included phenol/chloroform, DNA isolation kit, Cetyl Trimethyl Ammonium
Bromide
(CTAB) and Dodecyle Trimethyl Ammonium Bromide (DTAB).
Results:
Although DNA was
obtained from all of these methods, the phenol/chloroform and DNA
isolation kit
methods were found to be the most effective methods for obtaining high
quantity
DNA. RNA contamination was determined to be common in DTAB method. High
quantity
of DNA and RNA contamination in terms of quality was detected in CTAB
method.
Ruminant specific 16S rRNA primer was used to amplify genomic
DNA by
polymerase chain reaction and
all samples were
amplified except for some samples of DTAB.
KEYWORDS: CTAB, DTAB, Meat products, Phenol-Chloroform
INTRODUCTION
Meat is rich in proteins, vitamins, minerals such as iron, phosphor, zinc, and copper, which have a high biological value among the foods of animal origin; and also it is an appetizing, delicious and satisfactory food. Since meat includes exogen amino acids that cannot be synthesized by the body at an adequate and balanced amount, it has an important role in human nutrition. For this reason, meat or meat products must definitely be consumed by human beings (Arslan, 2013).
Sucuk, salami, and sausages are open to adulteration because of their production methods and the texture of the raw materials used. As the price of the meat and meat products increases, many adulterations may be observed in these products without considering the human health. Definition of the meat types used in meat products is important in terms of economic reasons, religious factors, the confirmation of the labels and preventing unjust competition (Sincer et al., 2010; İlhak and Güran, 2015).
It has been reported that the methods that are based on sensory qualities, anatomical differences, properties of the tissue and fat, histological properties, the amount of the glycogen in the meat are used in separation of meat and meat products. Also, some immunological, electrophoretic, serologic and genetic methods are reported to be used for this purpose (Hitchock and Crimes, 1985; Ekici and Akyüz, 2003; İlhak and Arslan, 2007a,b; Günşen et al., 2009).
In order to define the types of the meats with various molecular methods, which will be applied with genetic material; first of all, genomic DNA with a high molecular weight (of good quality and amount) must be obtained in a pure way. The DNA isolation method basically consists of three main successive stages which are- 1) Revealing DNA with high molecular weight with the lysis of the cell; 2) Separation of the DNA-protein complex,and having the DNA in soluble state using denaturation or proteolyses process; 3) Separation of the DNA from proteins, RNA and other macromolecules using simple enzymatic and/or chemical methods. The definition of the concentration of the DNAs that are obtained by using different methods is measured by using spectrophotometric methods that are based on absorption. The purity of the DNA molecule measured by spectrophotometer is obtained by rating the values obtained at 260 and 280 nm wavelengths (Topal Sarıkaya, 2004).
In recent years, identification of meat species in meat products obtained from various sales points of different cities by using different methods has become an important matter (Dalmasso et al., 2004; Özgen-Arun et al., 2014; Hou et al., 2015; Stamatis et al., 2015; Özşensoy and Şahin, 2016; Yin et al., 2016). For these studies, the first process is the isolation of the DNA, and it has been performed by using different methods such as DNA isolation kit (Özgen-Arun et al., 2014) and phenol/ chloroform (Krieg et al., 1983; Koh et al., 1998; İlhak and Arslan, 2007b; İlhak and Güran, 2015).
It has been reported in recent studies that DNA hybridization and PCR-based methods are used commonly for identification of meat species in meat products and examining the vegetable protein mixtures (Rahmati et al., 2016). For this reason, it is necessary that firstly, the purity of DNA with high concentration must be obtained in order to use the methods. The objective of our study was to compare four different DNA isolation methods in six different meat products.
MATERIALS AND METHODS
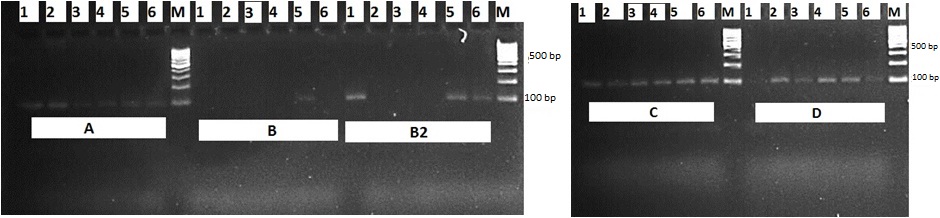
The material of the study consists of sucuk, salami, sausage, braised meat, meatball, and pastrami samples (Table 1). The DNA isolation was carried out in these six different meat products by using four different isolation methods. The DNAs obtained were amplified by using 104 base pair (bp)-long ruminant specific 16S rRNA primer (F: 5′-GAAAGGACAAGAGAAATAAGG-3′, R: 5′-TAGGCCCTTTTCTAGGGCA-3′) (Dalmasso et al., 2004) by polymerase chain reaction (PCR) and the usability of the DNAs was investigated.
DNA isolation methods: Small parts were taken from different points of the meat samples, and were mixed to homogenize. Then, one hundred mg of the samples were taken from this mixture, and the methods were performed. In the DTAB method, one hundred mg sample was taken and the analysis was performed. Another twenty mg sample was taken because the sample amount was in an excessive amount and there was no full-separation in the phases in the supernatant taking stage. The lower phase should also be taken when the supernatant is being taken in order to realize full separation of the phases and to take the supernatant with ease.
DNA isolation using DNA extraction kit: One hundred mg (A) from meat samples was taken. The gSYNCTM DNA Extraction Kit (Geneaid Biotech Ltd., Taiwan) was used for extraction of DNA from the samples. DNA isolation was performed according to manufacturer’s protocol. 100 mg meat sample was taken and transferred to a 1.5 mL micro centrifuge tube. Two hundred µL of GST Buffer and 20 µL of Proteinase K (20 mg/mL) were added and vortexed thoroughly, and then the tube was incubated at 60°C overnight. After incubation, the tube was centrifuged for two min at 16000xg and the supernatant was carefully transferred to a new 1.5 mL micro centrifuge tube. Two hundred µL of GSB Buffer was added to tubes and then shaken vigorously for 10 sec. Two hundred µL of absolute ethanol was added to the sample and mixed immediately by shaking vigorously for 10 sec. A GD Column was placed in 2 mL Collection Tube and all of the mixture (including any insoluble precipitate) was transferred to the GD Column. The tube was centrifuged at 16000xg for 1 min. Collection Tube was discarded then the GD Column was transferred to a new 2 mL Collection Tube. Four hundred µL of W1 Buffer was added to the GD Column and centrifuged at 16000xg for 30 sec then discarded the flow-through. The GD Column was placed back to Collection Tube. Six hundred µL of Wash Buffer was added to the GD Column and centrifuged at 16000xg for 30 sec and then discarded. The GD Column was placed back to Collection Tube and centrifuged again for 5 min at 16000xg to dry the column matrix. One hundred µL of pre-heated Elution Buffer was added into the center of the column matrix. The tube was kept at least 3 min to ensure that Elution Buffer is completely absorbed, and it was centrifuged at 16000xg for 30 sec to elute the purified DNA. The tube was stored along one day at +4°C. The DNA sample more than 100 ng/mL was diluted to a concentration of 100 ng/mL.
DNA isolation using DTAB method: One hundred mg (B1) and twenty mg (B2) from meat samples were taken and Dodecyl Trimethyl Ammonium Bromide (DTAB) method was performed by using isolation as previously mentioned (Kurar et al., 2012). Protocol procedure: 100 mg or 20 mg meat sample was taken and transferred to a 1.5 mL micro centrifuge tube. Eight hundred µL of Nuclear Lysis Buffer (12 g DTAB, 45 mL
DNA isolation using CTAB method: One hundred mg (C) from meat samples was taken and Cetyl Trimethyl Ammonium Bromide (CTAB) method was performed for extraction of DNA. Protocol procedure; 100 mg meat sample was taken and transferred to a 1.5 mL micro centrifuge tube. Two hundred and fifty µL of lysis buffer (0.25% SDS, 0.1 M EDTA pH 8.0) and 3 µL Proteinase K (10 mg/mL) were added, then vortexed thoroughly. The tube was incubated at 55°C for 20 min. Seventy-five µL of 3.5 M NaCl was added and mixed. Four-two µL 10% CTAB/0.7 M NaCl heated at 55°C was added, and mixed well by vortexing, and incubated at 65°C for 10 min. Four hundred µL of chloroform was added and vortexed. The tube was centrifuged at the highest setting (20000xg) for 5 min. The supernatant was transferred to a new micro centrifuge tube. Four hundred µL of phenol:chloroform:isoamyl alcohol (25:24:1) was added and mixed thoroughly up and down. The tube was centrifuged at 14000xg for 10 min at +4°C. The supernatant was transferred to a new micro centrifuge tube. Four hundred µL of 100% ethanol was added and mixed by shaking and incubated at room temperature for 5 min. The tube was centrifuged at 10 000 x rpm for 10 min at +4°C. The supernatant was poured and the step was repeated by adding 100% ethanol. The pellet was dried and fifty µL of TE was added. The tube was stored along one day at + 4°C. The DNA sample more than 100 ng/ml was diluted to a concentration of 100 ng/µL.
DNA isolation using phenol / chloroform method: One hundred mg (D) from meat samples was taken and one hundred µL of TE was added and then the sample was extracted by using a standard organic phenol/ chloroform method (Sambrook et al., 1989). The tube was stored along one day at +4°C. The DNA sample more than 100 ng/µL was diluted to a concentration of 100 ng/µL.
Absorbance definition of the DNA bands and the observing in the gel: In order to measure the purity levels of the DNA samples whose extractions were made, the optic densities of the samples at 260 nm and 280 nm wavelengths were measured by using a nanodrop spectro-photometer (mySPEC, VWR). The DNA samples were observed by using 0.6% agarose gel. Five µL DNA samples, 5 µL bidistillated water and 5 µL dye were mixed and loaded to gel. The DNA samples loaded to gel were separated at 100 V for 90 min. After this procedure was completed, the gel was observed with UV light in gel imaging system (Vilber Lourmat Quantum ST4).
Polymerase chain reaction method: Polymerase chain reaction (PCR) was carried out in 15 µL reaction volume including 1xMg++ free PCR buffer (Biolabs, NEB), 0.200 mM dNTPs (Biolabs, NEB),
The prepared PCR product was amplified using a touchdown PCR profile (Don et al., 1991) in thermal cycler (Bio Rad T-100) device. Touchdown PCR profile was used with two steps. The first step was initial denaturation at 95°C for 4 min, followed by 16 cycles of denaturation at 95°C for 30 sec, annealing beginning at 60°C and ending at 52°C for 30 sec and extension at 68°C for 30 sec. The annealing temperature was decreased 0.5°C per cycle until it reached 52°C. At the second step, 25 cycles of 95°C for 30 sec, 52°C for 30 sec and 68°C for 30 sec was applied. The final extension of 68°C for 5 min was applied in all reactions. The amplified PCR products were separated with 100 V for 60 min and loaded onto electrophoresis device (CBS Scientific) with 2% agarose gel and visualized on 365 nm UV.
RESULTS
In this study, the DNA isolation was carried out by using four different isolation methods in six different meat products (sucuk, sausage, salami, braised meat, meatball, and pastrami samples). DNA purity was measured by calculating the ratio of absorbance at 260 to 280 nm wavelengths, and the values were summarized in Table 1.
When the table is examined it is observed that generally high quality DNA was obtained at a high concentration with the isolation samples. It has been observed that the highest DNA concentration was found in samples, braised meat and pastrami, and in methods that phenol/chloroform and CTAB in spite of RNA contamination. It has also been determined that DNA with good quality and adequate concentration was obtained in the DNA isolation kit. It was determined that even in four different isolations (by using RNAse and at different amounts) in DTAB method,which was applied by using RNAse, there was RNA contamination. While the DNA amount obtained by DTAB method is relatively lower than other methods, obtained amounts are deemed adequate. Table 1 summarizes mathematical comparisons of obtained average DNA amounts by different methods. When all the methods are considered, it is observed that the best methods are phenol/chloroform and DNA isolation kit in terms of average DNA concentrations as well as their quality.
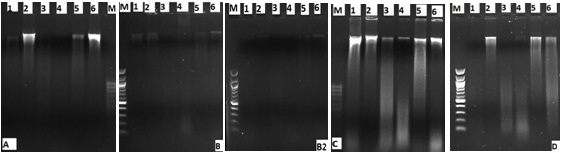
The DNA samples that were obtained by DNA isolation methods were observed by using 0.6% agarose gel (Figure 1). When a gel image was considered, it was observed that there were smears. These smears show that there are fractures in DNAs. The DNAs that were obtained from the meat products were amplified by using cattle specific 16S rRNA primers with PCR method (Figure 2). Some of the samples which were performed by two different amounts of DTAB method were not amplified. The PCR product was amplified in all samples obtained using the Kit, CTAB and phenol/chloroform methods.
Table
1. DNA
isolation methods, samples, DNA yield and OD values
| DNA isolation
Methods |
Samples |
ng / µL |
260/280 OD
values |
|
DNA Isolation
Kit |
Meatball |
37.578 |
1.750 |
| Salami |
71.958 |
1.799 |
|
| Sucuk |
51.053 |
1.787 |
|
| Braised meat |
154.241 |
2.077 |
|
| Sausage |
68.501 |
1.809 |
|
| Pastrami |
197.868 |
1.912 |
|
| DTAB Method
(B) |
Meatball |
67.266 |
2.340 |
| Salami |
35.711 |
2.430 |
|
| Sucuk |
27.476 |
2.099 |
|
| Braised meat |
76.361 |
1.730 |
|
| Sausage |
56.102 |
1.907 |
|
| Pastrami |
59.209 |
1.662 |
|
| DTAB Method
(B2) |
Meatball |
65.093 |
2.452 |
| Salami |
20.849 |
3.576 |
|
| Sucuk |
50.202 |
2.542 |
|
| Braised meat |
87.924 |
2.394 |
|
| Sausage |
60.612 |
1.999 |
|
| Pastrami |
130.274 |
1.611 |
|
| CTAB Method |
Meatball |
423.820 |
1.940 |
| Salami |
360.284 |
1.916 |
|
| Sucuk |
570.127 |
1.906 |
|
| Braised meat |
1457.560 |
2.228 |
|
| Sausage |
593.343 |
2.106 |
|
| Pastrami |
952.918 |
2.187 |
|
| Phenol/Chloroform
Method |
Meatball |
86.303 |
1.672 |
| Salami |
55.124 |
1.692 |
|
| Sucuk |
70.897 |
1.698 |
|
| Braised meat |
347.503 |
2.008 |
|
| Sausage |
92.557 |
2.000 |
|
| Pastrami |
338.529 |
1.418 |
DISCUSSION
It has been reported that taking one hundred mg sample will be adequate in molecular techniques worked on meat products (Özatay, 2012). One hundred mg meat sample was used as standard in all methods in this study. The PCR process was carried out with the DNA obtained as a result of the methods used, and adequate results was obtained.
It has been reported that two methods, which are CTAB as an organic method, and the commercial kit, are the most used methods in meat products. In CTAB method, concentration of DNA was high, and the quality was low (Pinto et al., 2007; Özatay, 2012). Similarly, it was deter-mined that although high concentration of DNA was obtained with the CTAB method in this study, there was RNA contamination.
It has also been reported that as a result of the two methods (DTAB and phenol/chloroform) used in rendering products obtained by exposing to high heat, DNA was obtained and amplified successfully using the PCR method (Kurar et al., 2012). Similarly, in this study, it was determined that all the samples that were obtained using the phenol/chloroform method, and many of the samples that were obtained using DTAB were amplified using PCR. Some of the samples which were performed by using two different amounts of DTAB method (one hundred and twenty mg) were not amplified. The reason for this was considered to be the DNAs obtained with DTAB were dissolved in high-amount buffer, and yet there was still unsolved residue in the tube; and for this reason, the PCR was not obtained in some of the samples.
The phenol/chloroform (Krieg et al., 1983; Koh et al., 1998; Matsunaga et al., 1999; Yetim et al., 2006; İlhak and Arslan 2007b; Kesmen et al., 2007; Kesmen et al., 2010; İlhak and Güran 2015) or DNA isolation kit (Kumar et al., 2011; Kesmen et al., 2012; Mane et al., 2012; Cawthorn et al., 2013; Ulca et al., 2013; Özgen-Arun et al., 2014; Ali et al., 2015; Stamatis et al., 2015; Safdar and Junejo, 2016; Yin et al., 2016) were used in studies, which were conducted for the purpose of isolating DNA and RNA from tissue; and mixtures of other substances were used for the identification of meat species. In this study, four different DNA isolation methods were tested, and DNAs were obtained. It was observed that when DNA yield and quality was considered, the best quality and purity was obtained in the phenol/chloroform method,which was used in previous studies as well, and with the DNA isolation kit. In the DTAB method, although DNA was obtained from meat products, it was not useful in isolation due to the RNA contamination. In addition, DNAs obtained from meat samples using DNA isolation loaded to gel electrophoresis, and obtained traces in the form of a smear in the samples. The reason for this was considered to be the fractures in the DNAs because they were exposed to heat.
CONCLUSION
It has been observed that DNA was obtained in the four different DNA isolation methods, and it may be used successfully in amplified PCR. It has also been observed that the best result was obtained with phenol/chloroform method among the four methods used, and we recommend that this method may be used successfully in meat products. For the researches who do not want to prefer the phenol/chloroform method because of its toxic effects, the second best method is the DNA isolation kit, which enables qualified and pure DNA isolation.CONFLICT OF INTERESTS
The authors declare that they have no conflict of interest.ACKNOWLEDGEMENT
This research was supported by the Scientific Research Project Fund of Cumhuriyet University under the project number V-017. The study has been presented as an abstract at the VI. National Veterinary Animal Science Congress, 1-4 June 2016, Cappadocia, Turkey.AUTHORS CONTRIBUTION
Both are contributed equally.REFERENCES
1. Ali ME, Razzak MA, Hamid SB, Rahman MM, Amin MA, Rashid NR, Asing (2015). Multiplex PCR assay for the detection of five meat species forbidden in Islamic foods. Food Chemistry, 177: 214-224. https:/doi.org/10.1016/j.foodchem.2014.12.0982. Arslan A (2013). Et muayenesi ve et ürünleri teknolojisi. İkinci baskı. Malatya, Medipress Yayıncılık; pp. 617-744.
3. Cawthorn DM, Steinman HA, Hoffman LC (2013). A high incidence of species substitution and mislabelling detected in meat products sold in South Africa. Food Control, 32: 440-449. https:/doi.org/10.1016/j.foodcont.2013.01.008
4. Dalmasso A, Fontanella E, Piatti P, Civera T, Rosati S, Bottero MT (2004). A multiplex PCR assay for the identification of animal species in feedstuffs. Molecular and Cellular Probes, 18: 81–87. https:/doi.org/10.1016/j.mcp.2003.09.006
5. Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS (1991). 'Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Research, 19: 4008. https:/doi.org/10.1093/nar/19.14.4008
6. Ekici K, Akyüz N (2003). Farklı hayvan türlerine ait çig etlerin SDS-PAGE yöntemiyle belirlenmesi üzerine bir araştırma. YYÜ Veteriner Fakültesi Dergisi, 14: 78-82.
7. Günşen U, Özcan A, Karaca MY, Kaygısız M (2009). Tüketime sunulan et ürünlerinde hile amaçlı yabancı et türü varlığının PCR yöntemi ile belirlenmesi. Bornova Veteriner Kontrol ve Araştırma Enstitüsü Dergisi, 31: 21-27.
8. Hitchock CH, Crimes AA (1985). Methodology for species identification. Meat Science, 15: 229-233.
9. Hou B, Meng X, Zhang L, Guo J, Li S, Jin H (2015). Development of a sensitive and specific multiplex PCR method for the simultaneous detection of chicken, duck and goose DNA in meat products. Meat Science, 101: 90-94. https:/doi.org/10.1016/j.meatsci.2014.11.007
10. İlhak Oİ, Arslan A (2007a). Rastgele çoğaltılmış polimorfik DNA yöntemiyle kanatlı etlerinde tür tayini. Fırat Üniversitesi Sağlık Bilimleri Dergisi, 21: 167-171.
11. İlhak Oİ, Arslan A (2007b). Identification of meat species by polymerase chain reaction (PCR)technique. Turkish Journal of Veterinary and Animal Sciences, 31: 159-163.
12. İlhak OI, Güran HŞ (2015). Authention of meat species in sucuk by multiplex PCR. İstanbul Üniversitesi Veteriner Fakültesi Dergisi, 41: 6-11.
13. Kesmen Z, Sahin F, Yetim H (2007). PCR assay for the identification of animal species in cooked sausages. Meat Science, 77: 649-653.
https:/doi.org/10.1016/j.meatsci.2007.05.018
14. Kesmen Z, Yetim H, Sahin F (2010). Identification of different meat species used in sucuk production by PCR assay. Gıda, 35: 81-87.
15. Kesmen Z, Yetiman AE, Şahin F, Yetim H (2012). Detection of chicken and turkey meat in meat mixtures by using real-time PCR assays. Journal of Food Science, 77: 167-173. https:/doi.org/10.1111/j.1750-3841.2011.02536.x
16. Koh MC, Lim CH, Chua SB, Chew ST, Phang STW (1998). Random amplified polymorphic DNA (RAPD) fingerprints for identification of red meat animal species. Meat Science, 48: 275-285. https:/doi.org/10.1016/S0309-1740(97)00104-6
17. Krieg P, Amtmann E, Sauer G (1983). The simultaneous extraction of high molecular weight DNA and RNA from solid tumours. Analytical Biochemistry, 134: 288-294. https:/doi.org/10.1016/0003-2697(83)90299-3
18. Kumar D, Singh SP, Singh R, Karabasanavar NS (2011). A highly specific PCR assay for identification of goat (Capra hircus) meat. Small Ruminant Research, 97: 76-78.
https:/doi.org/10.1016/j.smallrumres.2011.01.013
19. Kurar E, Özşensoy Y, Doğan MB, Nizamlıoğlu M (2012). DNA teknolojisi ile rendering ürünlerinde tür tespiti: Bir vaka raporu. Eurasian Journal of Veterinary Sciences, 28: 122-125.
20. Mane BG, Mendiratta SK, Tiwari AK (2012). Beef specific polymerase chain reaction assay for authentication of meat and meat products. Food Control, 28: 246-249. https:/doi.org/10.1016/j.foodcont.2012.05.031
21. Matsunaga T, Chikuni K, Tanabe R, Muroya S, Shibata K, Yamada J, Shinmura Y (1999). A quick and simple method for the identification of meat species and meat products by PCR assay. Meat Science, 51: 143-148. https:/doi.org/10.1016/S0309-1740(98)00112-0
22. Özatay Ş (2012). Moleküler metodların gıda kontrollerindeki uygulama alanları. Derleme, 5: 75-81.
23. Özgen-Arun Ö, Çiftçioğlu G, Altunatmaz SS, Atalay S, Savaşçı M, Eken HS (2014). Effect of processing on PCR detection of animal species in meat products. Kafkas Universitesi Veteriner Fakültesi Dergisi, 20: 945-950. https:/doi.org/10.9775/kvfd.2014.11428
24. Özşensoy Y, Şahin S (2016). Et ürünlerinde tür tayininin yapılmasında farklı yöntemlerin karşılaştırılması. Eurasian Journal of Veterinary Sciences, 32: 30-35. https:/doi.org/10.15312/EurasianJVetSci.2016115447
25. Pinto AD, Forte VT, Guastadisegni MC, Martino C, Schena FP, Tantillo G (2007). A comparison of DNA extraction methods for food analysis. Food Control, 18: 76-80. https:/doi.org/10.1016/j.foodcont.2005.08.011
26. Rahmati S, Julkapli NM, Yehye WA, Basirun WJ (2016). Identification of meat origin in food products-A review. Food Control, 68: 379-390.
https:/doi.org/10.1016/j.foodcont.2016.04.013
27. Safdar M, Junejo Y (2016). The development of a hexaplex-conventional PCR for identification of six animal and plant species in foodstuffs. Food Chemistry, 192: 745-749. https:/doi.org/10.1016/j.foodchem.2015.07.082
28. Sambrook J, Fritsch, EF, Maniatis T (1989). Molecular cloning: A laboratory manual. 2nd Edn., NY, USA: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; pp 9.16-9.19.
29. Sincer E, Şenyuva H, Gilbert J (2010). Et ve et ürünlerinde tağşiş ve orjinallik. Gıda&Yem Analiz'35 Dergisi, 7: 12-13.
30. Stamatis C, Sarri CA, Moutou KA, Argyrakoulis N, Galara I, Godosopoulos V, Kolovos M, Liakou C, Stasinou V, Mamuris Z (2015). What do we think we eat? Single tracing method across foodstuff of animal origin found in Greek market. Food Research International, 69: 151-155.
https:/doi.org/10.1016/j.foodres.2014.12.033
31. Topal Sarıkaya A (2004). DNA'nın izolasyonu ve analizi. In. Moleküler Biyolojide Kullanılan Yöntemler (Temizkan G, Arda N, ed). 2. Baskı. İstanbul: Nobel Tıp Kitapevleri; pp. 55-80.
32. Ulca P, Balta H, Çağın İ, Senyuva HZ (2013). Meat species identification and Halal authentication using PCR analysis of raw and cooked traditional Turkish foods. Meat Science, 94: 280-284. https:/doi.org/10.1016/j.meatsci.2013.03.008
33. Yetim H, Kesmen Z, Şahin F (2006). Kayseri ve Erzurum piyasasında satılan et ürünlerinde farklı hayvan türlerine ait etlerin PCR tekniği kullanılarak belirlenmesi üzerine bir araştırma. In: Türkiye 9. Gıda Kongresi, Bolu, Türkiye; pp 985-988.
34. Yin R, Sun Y, Yu S, Wang Y, Zhang M, Xu Y, Xue J, Xu N (2016). A validated strip-based lateral flow assay for the confirmation of sheep-specific PCR products for the authentication of meat. Food Control, 60: 146-150. https:/doi.org/10.1016/j.foodcont.2015.07.030